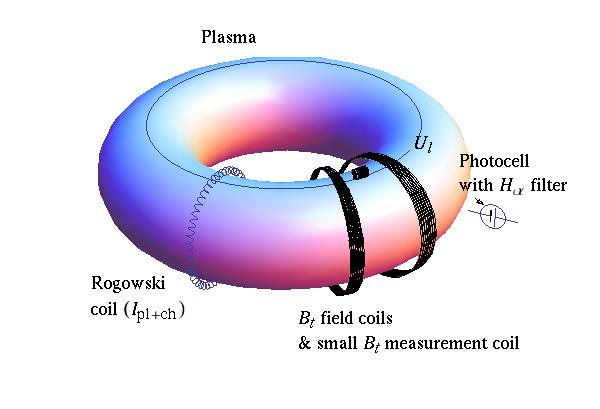
Tokamak GOLEM Basic diagnostics¶
Procedure (This notebook to download)
Prerequisities: function definitions¶
Load libraries
%matplotlib inline
import os
import numpy as np
import matplotlib.pyplot as plt
from scipy import constants, integrate, signal, interpolate
import sqlalchemy # high-level library for SQL in Python
import pandas as pd
For interactive web figures
import holoviews as hv
hv.extension('bokeh')
import hvplot.pandas
For conditional rich-text boxes
from IPython.display import Markdown
Define global constants.
base_URL = "http://golem.fjfi.cvut.cz/utils/data/{shot_no}/{identifier}" #remote access
data_URL = "http://golem.fjfi.cvut.cz/shots/{shot_no}/DASs/StandardDAS/{identifier}.csv" # TODO workaround
parameters_URL = 'http://golem.fjfi.cvut.cz/shots/{shot_no}/Production/Parameters/{identifier}'
destination=''
#os.makedirs(destination, exist_ok=True );
# try to get thot number form SHOT_NO envirnoment variable, otherwise use the specified one
shot_no = os.environ.get('SHOT_NO', 0)
The DataSource downloads and caches data (by full URL) in a temporary directory and makes them accessible as files.
ds = np.DataSource(destpath='/tmp')
def print_and_save(phys_quant, value, format_str='%.3f'):
print(phys_quant+" = %.5f" % value)
with open(destination+phys_quant, 'w') as f:
f.write(format_str % value)
update_db_current_shot(phys_quant,value)
def update_db_current_shot(field_name, value):
try:
engine = sqlalchemy.create_engine('postgresql://golem:rabijosille@192.168.2.116/golem_database')
except:
return
engine.execute(f"""UPDATE shots SET "{field_name}"={value} WHERE shot_no IN(SELECT max(shot_no) FROM shots)""")
def open_remote(shot_no, identifier, url_template=base_URL):
return ds.open(url_template.format(shot_no=shot_no, identifier=identifier))
def read_value(shot_no, identifier):
"""Return the value for given shot as a number if possible"""
value = open_remote(shot_no, identifier, base_URL).read()
return pd.to_numeric(value, errors='ignore')
def read_parameter(shot_no, identifier):
return open_remote(shot_no, identifier, parameters_URL).read().strip()
def read_signal(shot_no, identifier):
file = open_remote(shot_no, identifier, data_URL)
return pd.read_csv(file, names=['Time',identifier],
index_col='Time', squeeze=True) # squeeze makes simple 1-column signals a Series
def correct_inf(signal):
"""Inteprolate Inf values"""
signal = signal.replace([-np.inf, np.inf], np.nan).interpolate()
return signal
t_Bt = float(read_parameter(shot_no, 'TBt')) * 1e-6 # from us to s
t_CD = float(read_parameter(shot_no, 'Tcd')) * 1e-6 # from us to s
offset_sl = slice(None, min(t_Bt, t_CD) - 1e-4)
$U_l$ management¶
Check the data availability¶
loop_voltage = read_signal(shot_no, 'LoopVoltageCoil_raw')
polarity_CD = read_parameter(shot_no, 'CD_orientation')
if polarity_CD != 'CW': # TODO hardcoded for now!
loop_voltage *= -1 # make positive
loop_voltage = correct_inf(loop_voltage)
loop_voltage.loc[:t_CD] = 0
ax = loop_voltage.plot(grid=True)
ax.set(xlabel="Time [s]", ylabel="$U_l$ [V]", title="Loop voltage $U_l$ #{}".format(shot_no));
$B_t$ calculation¶
Check the data availability¶
It is as magnetic measurement, so the raw data only give $\frac{dB_t}{dt}$
dBt = read_signal(shot_no,'BtCoil_raw')
polarity_Bt = read_parameter(shot_no, 'Bt_orientation')
if polarity_Bt != 'CW': # TODO hardcoded for now!
dBt *= -1 # make positive
dBt = correct_inf(dBt)
dBt -= dBt.loc[offset_sl].mean()
ax = dBt.plot(grid=True)
ax.set(xlabel="Time [s]", ylabel="$dU_{B_t}/dt$ [V]", title="BtCoil_raw signal #{}".format(shot_no));
Integration (it is a magnetic diagnostic) & calibration¶
K_BtCoil = read_value(shot_no, 'K_BtCoil') # Get BtCoil calibration factor
print('BtCoil calibration factor K_BtCoil={} T/(Vs)'.format(K_BtCoil))
Bt = pd.Series(integrate.cumtrapz(dBt, x=dBt.index, initial=0) * K_BtCoil,
index=dBt.index, name='Bt')
ax = Bt.plot(grid=True)
ax.set(xlabel="Time [s]", ylabel="$B_t$ [T]", title="Toroidal magnetic field $B_t$ #{}".format(shot_no));
Plasma + Chamber current $I_{p+ch}$ calculation¶
Check the data availability¶
Because it is a magnetic measurement, the raw data only gives $\frac{dI_{p+ch}}{dt}$
dIpch = read_signal(shot_no, 'RogowskiCoil_raw')
if polarity_CD == 'CW': # TODO hardcoded for now!
dIpch *= -1 # make positive
dIpch = correct_inf(dIpch)
dIpch -= dIpch.loc[offset_sl].mean() # subtract offset
dIpch.loc[:t_CD] = 0
ax = dIpch.plot(grid=True)
ax.set(xlabel="Time [s]", ylabel="$dU_{I_{p+ch}}/dt$ [V]", title="RogowskiCoil_raw signal #{}".format(shot_no));
Integration (it is a magnetic diagnostics) & calibration¶
K_RogowskiCoil = read_value(shot_no, 'K_RogowskiCoil') # Get RogowskiCoil calibration factor
print('RogowskiCoil calibration factor K_RogowskiCoil={} A/(Vs)'.format(K_RogowskiCoil))
Ipch = pd.Series(integrate.cumtrapz(dIpch, x=dIpch.index, initial=0) * K_RogowskiCoil,
index=dIpch.index, name='Ipch')
ax = Ipch.plot(grid=True)
ax.set(xlabel="Time [s]", ylabel="$I_{p+ch}$ [A]", title="Total (plasma+chamber) current $I_{{p+ch}}$ #{}".format(shot_no));
Plasma current $I_{p}$ calculation¶
R_chamber = read_value(shot_no, 'R_chamber') # Get Chamber resistivity
print('Chamber resistivity R_chamber={} Ohm'.format(R_chamber))
L_chamber = 1e-6 # H
The chamber current $I_{ch}$ satisfies the equation (neglecting the mutual inductance with the plasma) $$U_l = R_{ch} I_{ch} + L_{ch} \frac{d I_{ch}}{dt}$$ Therefore, the following initial value problem must be solved to take into account the chamber inductance properly $$\frac{d I_{ch}}{dt} = \frac{1}{L_{ch}}\left( U_l - R_{ch} I_{ch}\right), \quad I_{ch}(t=0)=0$$
U_l_func = interpolate.interp1d(loop_voltage.index, loop_voltage) # 1D interpolator
def dIch_dt(t, Ich):
return (U_l_func(t) - R_chamber * Ich) / L_chamber
t_span = loop_voltage.index[[0, -1]]
solution = integrate.solve_ivp(dIch_dt, t_span, [0], t_eval=loop_voltage.index, )
Ich = pd.Series(solution.y[0], index=loop_voltage.index, name='Ich')
Ip_naive = Ipch - loop_voltage/R_chamber # creates a new Series
Ip = Ipch - Ich
Ip.name = 'Ip'
Ip_naive.plot(grid=True, label='naive $I_{ch}=U_l/R_{ch}$')
ax = Ip.plot(grid=True, label=r'using $U_l = R_{ch} I_{ch} - L_{ch} \frac{d I_{ch}}{dt}$')
ax.legend()
ax.set(xlabel="Time [s]", ylabel="$I_{p}$ [A]", title="Plasma current $I_{{p}}$ #{}".format(shot_no));
for I in [Ich.rename('$I_{ch}$'), Ipch.rename('$I_{ch}+I_p$'), Ip.rename('$I_p$')]:
ax = I.plot()
ax.legend()
ax.set(xlabel='Time [s]', ylabel='$I$ [A]', title='estimated plasma and chamber current and measured total')
plt.grid()
Plasma detect¶
The plasma detection is based on finding the time points of the greatest peaks in the first (smooth) derivative of $I_p$ which corresponds to the breaks in slope of $I_p$, i.e. the rise or fall to 0, respectively. The smooth derivative is found using the Savitzky-Golay filter.
# select oly time when CD is active to possibly skip Bt thyristor action
Ip_detect = Ip.loc[t_CD:]
dt = (Ip_detect.index[-1] - Ip_detect.index[0]) / (Ip_detect.index.size) # estimated sampling step
print('Sampling step {dt:.3g} s, sampling frequency {fs:.3g} Hz'.format(dt=dt, fs=1/dt))
window_length = int(0.5e-3/dt) # window fit over 1 ms
if window_length % 2 == 0: # must not be odd
window_length += 1
dIp = pd.Series(signal.savgol_filter(Ip_detect, window_length, polyorder=3, deriv=1, delta=dt),
name='dIp', index=Ip_detect.index) / 1e6 # [MA/s]
threshold = 0.5
plasma_start = dIp[dIp > dIp.max()*threshold].index[0] # plasma actually starts before the sharpest rise
plasma_end = dIp.idxmin() # due to inductance smoothing the greatest fall is the best predictor
is_plasma = int(plasma_start < plasma_end and dIp.abs().max() > 0.5)
print_and_save("b_discharge", is_plasma, "%.3f")
if not is_plasma:
plasma_start = plasma_end = np.nan
plasma_lifetime = 0
print_and_save("t_plasma_start", -1, "%.3f")
print_and_save("t_plasma_end", -1, "%.3f")
print_and_save("t_discharge_duration", -1, "%.3f")
else:
plasma_lifetime = plasma_end - plasma_start
print_and_save("t_plasma_start", plasma_start*1000, "%.3f")
print_and_save("t_plasma_end", plasma_end*1000, "%.3f")
print_and_save("t_discharge_duration", plasma_lifetime*1000, "%.3f")
fig, axs = plt.subplots(3, 1, sharex=True, dpi=200)
dIp.plot(grid=True, ax=axs[0])
axs[0].set(xlabel='', ylabel='$dI_p/dt$ [MA/s]', title='Plasma detection from $d I_p/dt$ in #{}'.format(shot_no))
Ip.plot(grid=True, ax=axs[1])
axs[1].set(ylabel="$I_{p}$ [A]")
loop_voltage.plot(grid=True, ax=axs[2])
axs[2].set(xlabel="Time [s]", ylabel="$U_{l}$ [V]")
for ax in axs:
for t in (plasma_start, plasma_end):
ax.axvline(t, color='k', linestyle='--', linewidth=2)
plt.savefig('icon-fig')
Overview graphs and parameters¶
For convenience time is saved and displayed in ms, currents in kA.
df_processed = pd.concat(
[loop_voltage.rename('U_loop'), Bt, Ip*1e-3, Ich*1e-3], axis='columns')
df_processed.index = df_processed.index * 1e3 # to ms
df_processed.head()
if is_plasma:
plasma_lines = hv.VLine(plasma_start*1e3) * hv.VLine(plasma_end*1e3)
else:
plasma_lines = hv.Curve([])
layout = df_processed['U_loop'].hvplot.line(ylabel='Uₗ [V]', xlabel='') * plasma_lines +\
df_processed['Bt'].hvplot.line(ylabel='Bₜ [T]', xlabel='') * plasma_lines +\
df_processed['Ich'].hvplot.line(label='Iᴄʜ') * df_processed['Ip'].hvplot.line(ylabel='Iᴄʜ, Iₚ [kA]', label='Iₚ', xlabel='time [ms]') *\
plasma_lines
plot = layout.cols(1).opts(
hv.opts.Curve(width=600, height=200, title='', ylim=(0, None), show_grid=True),
hv.opts.VLine(color='black', alpha=0.7, line_dash='dashed')
)
hvplot.save(plot, 'homepage_figure.html')
plot
fname = 'basig_diagnostics_processed.csv'
df_processed.to_csv(fname)
Markdown("[Time series in graph in CSV format](./{})".format(fname))
units = ['V', 'T', 'kA', 'kA']
plasma_start_ms = plasma_start *1e3
plasma_end_ms = plasma_end *1e3
plasma_lifetime_ms = plasma_lifetime * 1e3
if is_plasma:
heading = Markdown("### Basic diagnostics values during plasma\n\n"
"plasma lifetime of {:.3g} ms, from {:.3g} ms to {:.3g} ms".format(
plasma_lifetime_ms, plasma_start_ms, plasma_end_ms))
else:
heading = Markdown("### No plasma detected (vacuum discharge)")
heading
if is_plasma:
plasma_sl = slice(plasma_start_ms, plasma_end_ms)
else:
plasma_sl = slice(0) # TODO really use whole discharge ?
df_during_plasma = df_processed.loc[plasma_sl]
df_overview = df_during_plasma.quantile([0.01, 0.5, 0.99]) # use quantiles to skip peaks
df_overview.index = ['min', 'mean', 'max'] # actually quantiles, but easier to understand
if is_plasma:
df_overview.loc['start'] = df_during_plasma.iloc[0]
df_overview.loc['end'] = df_during_plasma.iloc[-1]
else:
df_overview.loc['start'] = df_overview.loc['end'] = np.nan
df_overview.loc['units'] = units
# make units row first
df_overview = df_overview.iloc[np.roll(np.arange(df_overview.shape[0]), 1)]
df_overview
for agg in ('mean', 'max'):
for quantity, value in df_overview.loc[agg].iteritems():
print_and_save(quantity+'_'+agg, value)
End of overview row