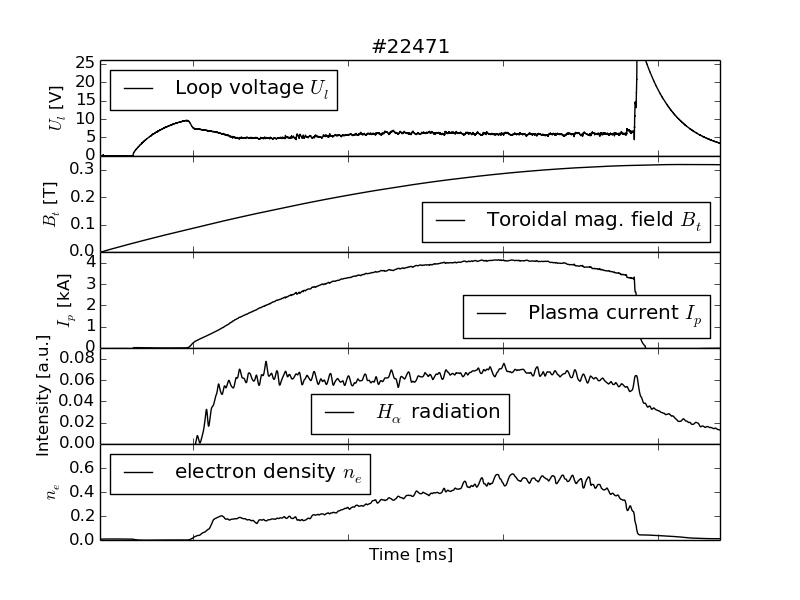
import numpy as np
import matplotlib.pyplot as plt
shot_no = 22471
identifier = "loop_voltage"
# create data cache in the 'golem_cache' folder
ds = np.DataSource('golem_cache')
#Create a path to data and download and open the file
base_url = "http://golem.fjfi.cvut.cz/utils/data/"
def get_data(identifier):
data_file = ds.open(base_url+ str(shot_no) + '/' + identifier)
return np.loadtxt(data_file)
fig = plt.figure(1)
plt.subplots_adjust(hspace=0.001)
sbp1=plt.subplot(511)
data1 = get_data('loop_voltage')
plt.ylim(0,26)
plt.yticks(np.arange(0, 30, 5))
plt.title('#'+str(shot_no))
plt.ylabel('$U_l$ [V]')
plt.plot(data1[:,0]*1000, data1[:,1], 'k-',label='Loop voltage $U_l$' )
plt.legend(loc=0)
sbp1=plt.subplot(512, sharex=sbp1)
data2 = get_data('toroidal_field')
plt.yticks(np.arange(0, 0.5 , 0.1))
plt.ylim(0,0.35)
plt.ylabel('$B_t$ [T]')
plt.plot(data2[:,0]*1000, data2[:,1], 'k-',label='Toroidal mag. field $B_t$')
plt.legend(loc=0)
sbp1=plt.subplot(513, sharex=sbp1)
data3 = get_data('plasma_current')
plt.yticks(np.arange(0, 4.5, 1))
plt.ylim(0,4.5)
plt.ylabel('$I_p$ [kA]')
plt.plot(data3[:,0]*1000, data3[:,1]/1000, 'k-',label='Plasma current $I_p$')
plt.legend(loc=0)
sbp1=plt.subplot(514, sharex=sbp1)
data4 = get_data('photodiode_alpha')
plt.yticks(np.arange(0, 0.09 , 0.02))
plt.ylim(0,0.09)
plt.ylabel('Intensity [a.u.]')
plt.plot(data4[:,0]*1000, data4[:,1], 'k-',label='$H_\\alpha$ radiation')
plt.legend(loc=0)
sbp1=plt.subplot(515, sharex=sbp1)
data5 = get_data('electron_density')
plt.yticks(np.arange(0, 0.8 , 0.2))
plt.ylim(0,0.8)
plt.ylabel('$n_e$')
plt.xticks(np.arange(8, 30, 5))
plt.xlim(5,25)
plt.xlabel('Time [ms]')
plt.plot(data5[:,0]*1000, data5[:,1]*1e-19, 'k-',label='electron density $n_e$')
plt.legend(loc=0)
xticklabels = sbp1.get_xticklabels()
plt.setp(xticklabels, visible=False)
plt.savefig('basicgraph.pdf')
plt.savefig('basicgraph.jpg')
plt.show()
|
|